DNA Repair
DNA Damage
Nitrous acid
Exposure to nitrous acid can accelerate spontaneous deamination of bases. They can result in the following:
- Cytosine → Uracil
- 6-methylcytosine (methylated cytosine) → Thymine
- Adenine → Hypoxanthine
- Guanine → Xanthine
UV radiation
Leads to the formation of free radicals, which can cause the double bonds in pyrimidines to link, forming a dimer. Thymine bases are particularly susceptible to this.
Mutation Types
Silent
Nucleotide mutation encoding for the same amino acid as the original sequences. Does not affect phenotype.
Transition
Point mutation involving a purine to a purine or a pyrimidine to a pyrimidine
Transversion
Point mutation involving a purine to a pyrimidine or a pyrimidine to a purine
Depurination
Removal of a purine nitrogenous base through hydrolysis
Methylation
Addition of a methyl group through exposure to an alkylating agent. This can be purposeful and beneficial.
Cytosine is a mutation hotspot
Cytosine-guanine dinucleotides are often methylated. When 5-methylcytosine is deaminated, the resulting thymine will result in an adenine rather than a guanine being placed.
This issue can sometimes be resolved as some repair systems will preferentially replace a T with a C when encountering a G/T mismatch, but this isn't a robust solution.
C-G sequences are generally palindromes and can be recognized as a hotspot for mutations.
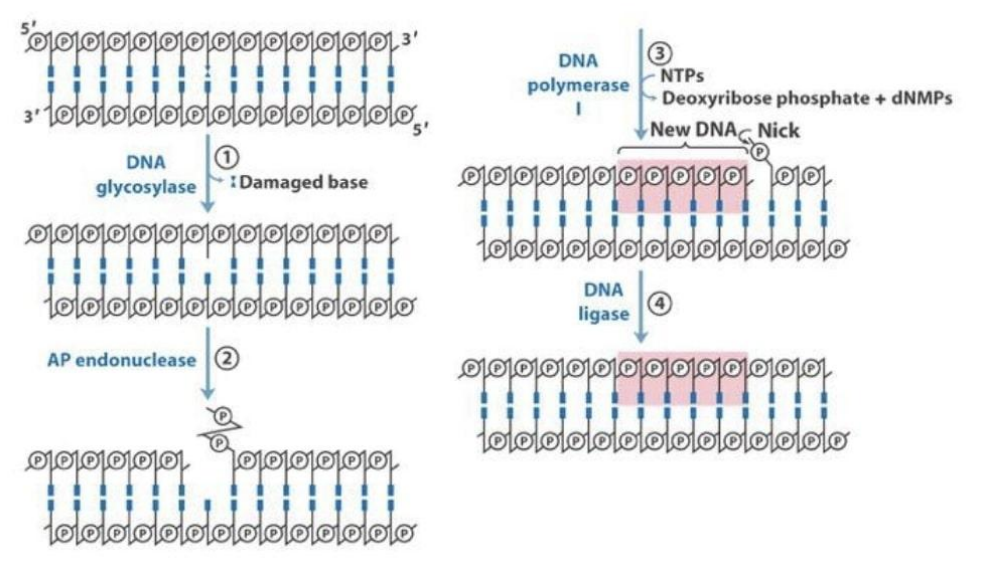
Base Excision Repair
Fixes small types of damage like deamination or depurination.
- DNA glycosylase: Removes damaged base leaving a blank sugar moiety
- AP endonuclease: Removes the sugar moiety
- DNA polymerase I: Replaces the damaged base using the template strand
- DNA ligase: Seals the strand closed (closes the "nick")
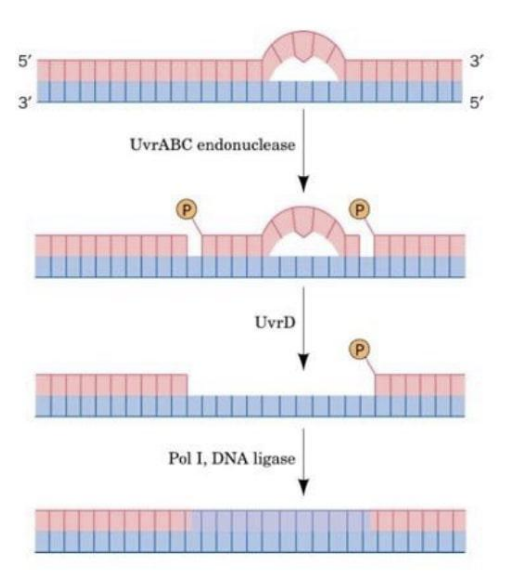
Nucleotide Excision Repair
Fixes base mutations like pyrimidine dimers.
- UvrABC endonuclease: Creates a "nick" on either side of mutation
- UvrD helicase: Removes the improper stretch of DNA
- DNA polymerase I: Replaces the missing DNA using template strand
- DNA ligase: Seals the strand closed
Clinical Pearls
Hereditary Nonpolyposis Colorectal Cancer (HNPCC)
Inability to introduce nicks and therefore properly repair mismatch mutations. Results from mutations in hMLH1 or hMSH2 genes.
Xeroderma Pigmentosum (XP)
Hypersensitivity to UV light and therefore predisposed to skin cancer. Results from a mutation in Transcription Factor H, which exhibits helicase activity. This makes it hard to fix thymine dimers with nucleotide excision repair.
Cockayne's Syndrome & Trichothiodystrophy
Will present with abnormal growth and neurological symptoms. Also involves a Transcription Factor H mutation as in XP, however these mutations will affect Transcription-Coupled Repair rather than nucleotide excision repair.