Homologous Recombination
The process by which homologous chromosomes exchange alleles.
Mechanism
In prophase I, homologous chromosomes are brought in proximity. During this time, portions of the chromosomes cross over forming chiasmata. These sections will recombine and swap before separating again into each daughter cell.
Functions
- Restarting DNA replication forks (when stalled)
- Ensure proper segregation of homologous chromosomes (nondisjunction)
- Genetic diversity (mixing of alleles)
- Unlinks alleles normally located near each other (and would otherwise be transcribed together)
Clinical pearl: Down syndrome
Non-disjunction will lead to chromosomal abnormalities, including trisomy which is present in Down Syndrome.
Repairing Double Strand Breaks
Homologous recombination
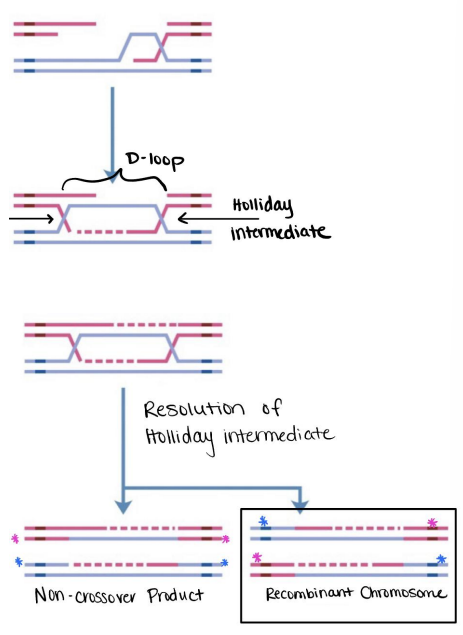
When a strand separates, exonucleases will degrade the DNA on the parent chromosome. The 5' ends will be degraded more than the 3' ends, exposing more of the 3' single-stranded DNA.
The 3' end will bind RAD51 which searches for homology along the intact homologous chromosome from the other parent. It will facilitate strand invasion, creating D-loop. The D-loop is expanded via DNA polymerase using the intact parent strand as a template.
As the strand is expanded, it will form two crossover points called Holliday intermediates (or junctions). These will either undergo recombination or form a non-crossover product which occurs when the two ends of the chromosomes come from the same parent, and is therefore not a true recombination.
Advantages
- Accurate
- Seamless continuation of replication
Disadvantages
- Can only be used in the S and G2 phase
Non-homologous end joining
Gluing the broken pieces of DNA back together. If this sounds janky, you would be correct.
- Ku heterodimer proteins bridge between the two pieces
- Exonucleases shave off the ends until they're compatible
- Ligases anneal the strands back together
Advantages
- Can also be used in the G1 phase (without homologous chromosomes)
Disadvantages
- Error prone repair
Restarting Replication Fork
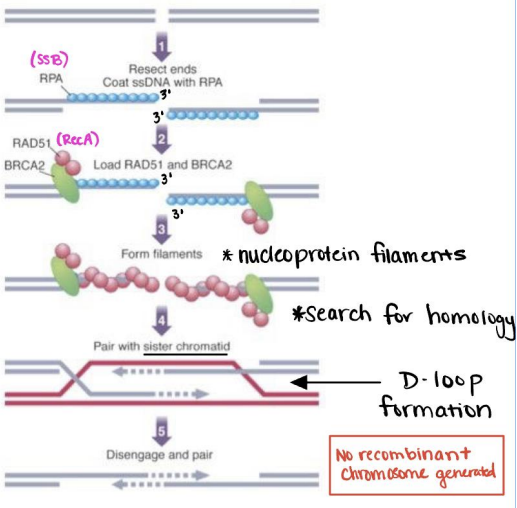
Replication will stall if a replication fork encounters a nick on a leading strand and will create a double strand break. This looks a lot like recombinant correction of double strand breaks, but won't generate a recombinant product as it uses the sister chromatid as a template which should be identical.
No recombinant product is important as the generic replication fork occurs in non-meiotic processes which should be generating exact copies, not facilitating diversity like homologous recombination.
- Exonuclease degrades the exposed ends
- RPA coats the exposed 3' single stranded DNA in RPA protein
- RAD51 binds BRCA2 and RPA protein and forms nucleoprotein filaments
- The nucleoprotein filaments search for homology against the sister chromatid (which should be an exact copy)
- RAD51 initiates strand invasion and D-loop formation
- DNA polymerase expands D-loop using the other chromosome as a template
Methods for Bypassing DNA Lesions
Forking and merging (Yes, like GitHub)
Thymine dimers will block high-fidelity DNA polymerase. However, because the replication fork has separated the strands there's no available template.
However, the lagging strand can continue as it presumably does not have the dimer. It will replicate the sequences past the point of the dimer and then will reanneal the correct DNA strands via RAD51. This creates a fork and allows the lagging strand to be used as a template.
As the strands are now back together, nucleotide excision repair can fix the dimer. This process allows replication to continue before the dimer is repaired.
Using Error-prone DNA Polymerase
Error-prone DNA polymerase will be swapped in. These contain looser active sites that more readily accept nucleotides, even if the DNA is distorted. These will only be active for short stretches of DNA and will be replaced with the high-fidelity DNA polymerase after the repair.
Overuse of these can create mutations.
Clinical pearl: Xeroderma Pigmentosum (XP)
Sensitivity to UV light which can be treated by knocking out a specific error-prone DNA polymerase. In healthy individuals, lacking this error-prone DNA polymerase can create cancers.
For more detail, see here: DNA Repair > Xeroderma Pigmentosum (XP)
BRCA Mutations
BRCA1 and BRCA2 aid in DNA repair.
BRCA1: Mediator in the protein kinase cascade response to DNA damage. Is relevant in transcription, DNA repair, senescence, and apoptosis
BRCA2: Is a docking site for RAD51 (see homologous recombination repairs)
Mutations that knockout/inactivate the BRCA1 and/or BRCA2 genes eliminate the option to carry out homologous recombination. Therefore, the non-recombinant end joining pathway must be utilized, which makes mutations more likely.
NOTE: BRCA gene mutations are linked to Breast Cancer, hence the naming of the gene (BReast CAncer)