Immune Diversity And Recognition Of Antigens
Major Histocompatibility Complex (MHC)
Expressed on cell surfaces with peptide binding clefts which hold antigens. Clefts have variability to attach many different antigens. They are not stable without an antigen and require a chaperone protein when generated. Once the presenting peptide is loaded in the ER, it immediately travels to the cell surface.
MHC I
On all nucleated cells. Presents internally-made peptides to CD8 to kill virally infected cells. Peptides are 8-10 amino acids long.
MHC II
On all antigen presenting cells (macrophages, dendritic cells, B cells, thymic epithelial cells). Presents externally made peptides to CD4 cells which activate the adaptive immune system. Peptides are 13-17 amino acids long (variable length).
Cross presentation
Dendritic cells present endocytosed, exogenous viral proteins on MHC I. This allows them to activate CD8 cells.
Polymorphism and Polygeny
Polymorphism is when multiple alleles for the same gene are present in a population. Polygeny is when each person has multiple copies of the gene.
There are multiple alleles for MHC and everyone has multiple combinations of the alleles, allowing lots of variability for antigen binding.
Transition to Adaptive Immunity
First, the Innate Immunity and the Complement System recognizes and begins a defense.
Secondarily, dendritic cells endocytose foreign bodies and present them to T cells. Activated T cells release cytokines and signal B cells to differentiate into plasma cells, releasing antibodies specific to the pathogen.
T/B cell receptors
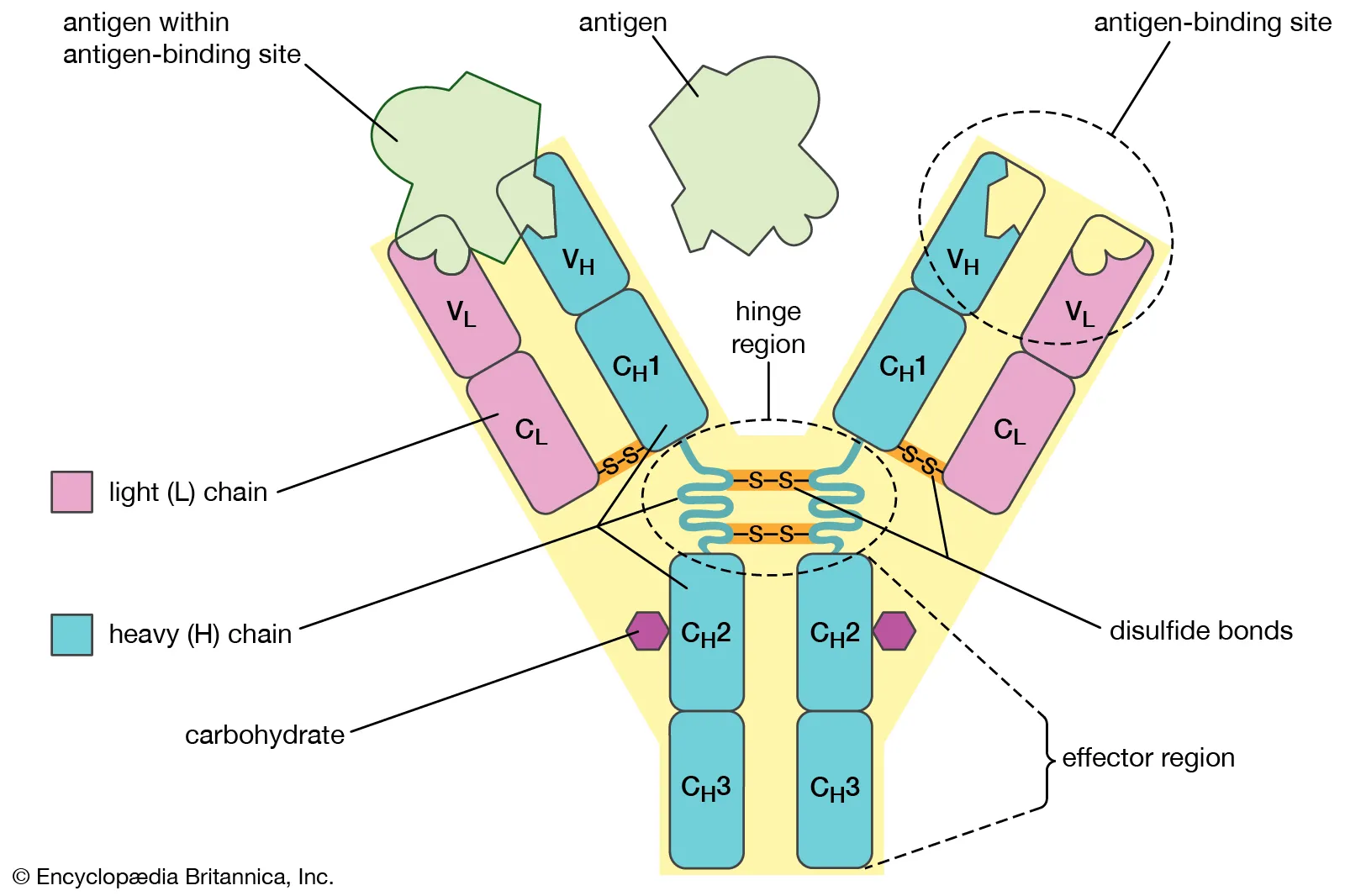
B cell receptors (BCRs)
Antibodies which are secreted from plasma cells. They are made of 4 polypeptide chains, 2 heavy and 2 light. Each antibody either has a kappa light chain or a lambda light chain. This gives the antibody two binding sites.
The constant region is the same on each B cell, called the Fc region. The variable region is different for each antibody and contains the antigen binding site.
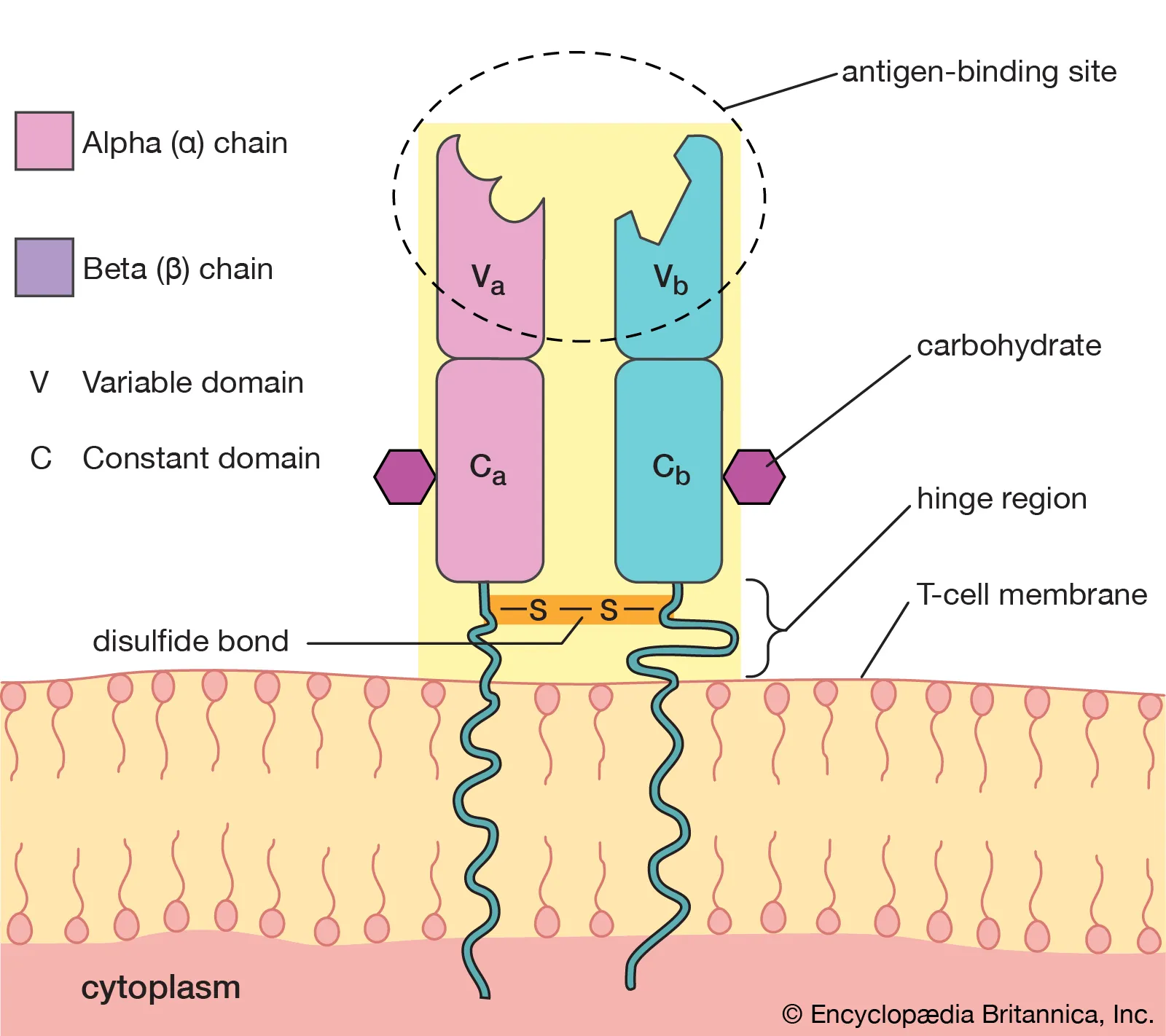
T cell receptors (TCRs)
Only on the cell surface of T cells. Made of 2 polypeptide chains: Either alpha/beta or gamma/delta. These are also heavy/light chains.
The constant region is the same on each T cell. The variable region is different for each T cell and contains the antigen binding site.
Valency
The number of antigen binding sites a receptor has.
B cell valency is at least 2 as it has two copies of the heavy/light chains and two binding sites.
T cell valency is always 1 as it has 1 binding side.
Generation of Diversity
V(D)J recombination
Main method used to make immune receptors. Mechanism is the mixing of gene segments and imprecise joining of recombined segments.
Receptors are made of Variable (V), Joining (J), and Diversity (D) segments. These are coding sequences for the receptor. The Constant (C) region joins the party after transcription.
Light chain assembly (V + J)
- V combines with J and DNA between them is deleted
- Primary RNA transcript is made
- C region is added via RNA splicing
Heavy chain assembly (D + J + V)
- D combines with J and DNA between them is deleted
- DJ combined with V and DNA between them is deleted
- C region is added via RNA splicing
Recombination signal sequences (RSSs) surround the coding segments. RSSs have different spacer lengths which tags which type of segment (V, J, or D) it is for recombination. These follow the 12/23 base pair length rule, which insures a 12 bp tagged RSS segment will be paired with a (different) 23 bp tagged RSS segment.
Recombination steps
V(D)J recombinase is made of RAG1 and RAG2 (Recombination Activating Gene).
- RAG1/2 will recognize the 12/23 RSS combination and create a double stranded DNA break between RSS pairs
- RAG1/2 removes the RSSs, creating hairpins (phosphodiester bond linking the two DNA strands at the end) on the ends of the coding sequences
- Signal ends are joined via non-homologous end joining
- Hairpins are opened using the Artemis protein
- Random nucleotides are added to the DNA non-specifically using terminal deoxytransferase. Exonucleases can also be used to remove nucleotides here
- Coding ends are joined by non-homologous end joining
Random combination of antigen heavy/light chains
Heavy/light chains undergo recombination independently, then combine to create diversity.
Somatic Hypermutation
The B cell rapidly divides to produce new variations in its antibodies. AID converts C to U in hypervariable regions.
Mechanisms of mutation
- U residues are removed by Base Excision Repair (BER) proteins and DNA glycosylase. The openings sites are repaired by translation synthesis (insertion of a random base).
- Mismatch Repair (MMR) proteins cleave DNA segments around the U error and DNA polymerase replaces it, with a preference for A-T.
- Mismatches which are missed by the above mechanisms result in two different daughter strands, increasing diversity.
Clinical Pearl: SCID
Mutations that inactivate the genes encoding RAG1/2 cause a form of SCID in which the patient does not have B or T cells, only NK cells. Partial inactivation can lead to variable immunodeficiency or Omenn syndrome (complete absence of B cells and low/normal T cells, with reduced T cell diversity).
Causes
- Mutations in several genes
- IL-2 gC deficiency: X-linked SCID (most common)
- ADA deficiency
- Interleukin receptor mutation (RAG1/2, JAK3, etc.)
Germline Configurations
Antibodies
The kappa light chain has 35-40 V segments and 5 J segments.
The lambda light chain has 30 V segments and 4-5 J segments.
The heavy chain has V(D)J segments.
T cell receptor
The most common TCR configuration is alpha/beta. The alpha chain has V/J segments while the beta chain has V/D/J segments.
The gamma/delta configuration has V/D/J segments. Delta is actually embedded in the alpha chain. If the alpha locus undergoes recombination, the delta locus gets deleted.
Selection
Signaling leading to cell death.
B cells
Occurs in the bone marrow. Pro-B cells undergo V(D)J recombination. At this point, the heavy chain and a surrogate light chain are expressed. The cell then undergoes positive selection, in which the cell must receive a signal through its new receptor to survive. If selected, it will recombine and express the light chain. If it is not selected, the other heavy chain allele can also attempt V(D)J recombination.
Next, negative selection. If the receptor binds to neighboring B cells, it means it was cross-linked by a self-antigen and any signal will cause the cell to die or be recombined.
T cells
Occurs in the thymus. The T cell must bind to an antigen presented by MHC on a thymic cell to survive. It must also bind to the self-antigen and self-MHC with moderate affinity, leading to positive selection. If it binds too strongly, it undergoes negative selection to control for over-reactivity.
If it binds to MHC II, it becomes a CD4 cell. If it binds to MHC I, it becomes a CD8 cell.
Developmental stages
- Double negative: Neither CD4/CD8 expressed
- Double positive: Both CD4/CD8 expressed
- Single positive: Either CD4/CD8 expressed
Antigen Binding
All antibodies and TCRs have a CDR (complementary determining region) which are super diverse loops at the tips of the variable region which bind the antigen. There are at least 6 CDRs for every 1 antigen domain. The antigen/antibody interaction is based on weak molecular interactions.
Epitope: The part of antigen which binds the antigen receptor
Paratope: The part of the receptor which interacts with the epitope
Idiotope: The part of the receptor that includes the paratope
Guanyltope: The part of the... just kidding
Linear epitopes are straight peptide sequences which are intact even when the antigen is denatured. A conformational epitope is created by folding the antigen and thus is buggered when denatured.
Differences in antibody and TCR binding
| Antibody | TCR | |
|---|---|---|
| Valence | Multivalent (2+ binding sites) | Monovalent |
| Epitope | Linear and conformational | Linear only |
| Binding targets | Everywhere | MHC presented antigens |
| Molecular target | Any macromolecule | Peptides |
TCR/MHC/Peptide complex
T cells are activated by MHC I/II antigens, but require additional co-stimulatory signals. Cells which do not receive this signals become anergic (inactive), can die (creating antigen tolerance), or become T regulatory cells.
CD4 (on helper T cells) and CD8 (on killer T cells) are coreceptors which stabilize the MHC complex on the T cell. This further restricts CD4 to MHC II and CD8 to MHC I.
All T cells have CD3 which is required to transduce signals upon antigen binding. The CD3 portion has epsilon, gamma, and delta chains as well as an internal zeta chain. These each have immunoreceptor tyrosine activation motifs (ITAMs) which transduce signals through tyrosine phosphorylation upon antigen binding.